Somnolink – a national initiative to improve data sharing for better healthcare and research in obstructive sleep apnea
Dagmar Krefting 1,2Michael Arzt 3
Moritz Brandt 4
Miriam Goldammer 4
Franz Ehrlich 4
Joachim T. Maurer 5
Thomas Penzel 6
Tony Sehr 4
Andrea Rodenbeck 7
Philip Zaschke 1
Christoph Schöbel 8
1 University Medical Center Göttingen, Göttingen, Germany
2 Campus Institute Data Science, Georg-August University, Göttingen, Germany
3 Department of Internal Medicine II, University Hospital Regensburg, Germany
4 Faculty of Medicine and University Hospital Carl Gustav Carus, TUD Dresden University of Technology, Dresden, Germany
5 University Hospital Mannheim, Ruprecht-Karls-University Heidelberg, Germany
6 Charite – Universitätsmedizin Berlin, Berlin, Germany
7 Evangelical Hospital Goettingen-Weende, Göttingen, Germany
8 University Medicine Essen, Essen, Germany
Abstract
Introduction: Obstructive sleep apnea (OSA) has a high prevalence and is associated with several severe health conditions. However, it might remain undetected and even when diagnosed, patients show relatively low adherence to the standard therapy.
Methods: To optimize OSA diagnosis and therapy, the project Somnolink addresses relevant improvement targets along the patient path by leveraging existing and upcoming medical informatics methods, ranging from health data integration to ML-based clinical decision support. We identified main improvement targets, the relevant data types, data items and data exchange methods, and analysed to which extent health standards and solutions exist already.
Results: The different targets require partly similar, partly diverging data types, from questionnaires to multidimensional biosignal recordings.
Conclusion: Sleep medicine is a multidisciplinary specialty and healthcare standards typically address only parts of the full data spectrum, including technical standards for biosignal recordings. However, most of the relevant data items and measures are covered by one of the semantic standards, allowing for semantic interoperability.
Keywords
sleep medicine, health information interoperability, data management, computer-assisted decision making
Introduction
Sleep is continuously gaining interest as a major health factor [1]. Among the numerous sleep disorders, obstructive sleep apnea (OSA) – the intermittent cessation of breathing during sleep due to an obstructed upper airway – is the most prevalent disorder in clinical sleep laboratories. An estimated 936 million people between 30 and 69 years worldwide have more than 5 respiratory events per hour while sleeping, each event lasting for at least 10 seconds. More than 40% of them have moderate to severe OSA [2]. OSA may cause symptoms such as excessive daytime sleepiness, repeated awakening reactions, dry mouth, or headache, or may be present without symptoms. OSA is associated with many cardiovascular, cerebrovascular, and metabolic diseases, as well as with cognitive deficits, cancer and other sleep disorders [3].
The diagnostic path typically differs – depending of the healthcare system – but typically encompasses screening, functional status and Quality of Life questionnaires, symptom assessment, physical examination, and may include medical imaging. However, for assessment of the diagnostic criteria, overnight sleep recordings of physiological signals are required. Depending on the country, home sleep testing (HST) recording breathing, airflow, oxygen saturation and electrocardiography (ECG), or recordings including electroencephalography (EEG) of up to 40 physiological signals, so-called polysomnographies (PSG), are performed. The latter is considered as gold-standard for the assessment of sleep stages and sleep disorders, but is typically more expensive than HST. For example, in Germany, PSG for diagnosis is only reimbursed by the statutory health insurances if the patient has comorbidities or screening HST did not deliver clear results. The standard therapy is a breathing mask providing continuous positive air pressure (CPAP). However, many patients abort CPAP therapy, up to 50% refuse CPAP therapy within one year [4]. Other individual treatment options are mandibular repositioning devices, positional therapy, surgical treatment, and hypoglossal nerve stimulation.
Targets of improved OSA health care and data-driven approaches
Due to the high prevalence, the strong association with many severe health conditions, the relatively low adherence to the standard therapy, and a low rate of the use of secondline therapies in patients who quit standard therapy, sleep experts see an urgent need to improve OSA health care. This includes (a) identification of OSA patients with high risk for severe health conditions including the reduction of the current gender bias, (b) optimization of the diagnostic path, (c) individualized therapy, and (d) improvement of therapy adherence.
The variety of data types and sources in OSA diagnosis and therapy, including longterm biosignal recordings, offers high potential for reaching these goals by digitalization, health data sharing, and ML-based decision support systems. While legal, ethical, organizational and technical challenges still hinder full exploitation of these benefits, substantial improvements in all areas have been made in the last years, in particular in Germany, with a still low degree of digital transformation in healthcare. For example, the German Medical Informatics Initiative (MII), set up common organizational, ethical and legal frameworks for health data sharing among all 36 university medical centers, with the first non-university hospitals already joining the initiative [5]. The German Health Data Utilization Act [6] and the Act to accelerate the digitalization of the healthcare system [7], which have both come into force in March 2024, aim for better use of the electronic health record and other digital health data for a learning healthcare system. Advancements in health data standardization such as the international patient summary or the MII’s core data set (MII-CDS) for inpatient healthcare data are a good base for structured and semantically annotated data relevant for OSA.
The Somnolink project within the framework of the MII funding scheme aims to address the above mentioned targets in an interdisciplinary and patient centered approach.
Six University Medical Centers (UMCs), a local hospital with a stationary sleep lab and an academic computer center (GWDG) have joined clinical and medical informatics expertise, accompanied by self-help groups and outpatient sleep labs. In this article, we introduce the goals, the medical informatics and data-science based solution approaches, the so-far reached results, the identified challenges and further steps. While Somnolink focuses clinically on OSA, many of the methods may be easily adopted to other sleep disorders, but also to other complex medical diagnostic and therapeutic paths.
Methods
We related the improvement targets and the typically acquired health data to the different phases of the patient path in OSA, as depicted in Figure 1 [Fig. 1]. Based on this schema, we propose data-driven solution approaches, where methods of medical informatics – including ML-based decision support systems – can contribute to the targets. The identification of the relevant data sources is based on the practice of the clinical partners. The patient path is based on the German Guidelines for OSA diagnostics, that follows a 4-step approach, from the assessment of the pretest probability via anamnesis and questionnaires (step 1) and physical examination (step 2) in a family practice, sleep diagnostics by an outpatient sleep specialist (step 3), and comprehensive diagnostics in a sleep lab (step 4) [8] .
Figure 1: Overview over the project’s topics and goals.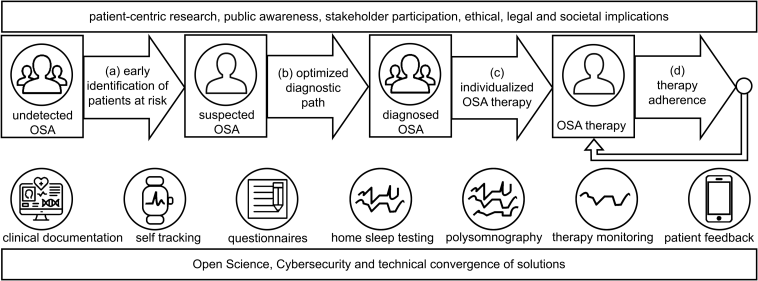
The rectangles with the person icons depict the different phases in a typical OSA patient path. The improvement targets are depicted by the arrows between the phases, as they all aim to optimize the transitions between the phases. The icons below the patient path depict the different data sources and types. They are roughly ordered along the path, but typically cannot be assigned to a single phase. For example, questionnaires might be employed in all phases but are mostly used in screening and diagnosis. The rectangles above and below show the overarching topics relevant for all targets and phases.
Standards for these data types are identified by inspection of common health standards (DICOM, IEEE 11073, EDF, HL7 FHIR basic resources and publicly available profiles, SNOMED CT, LOINC, ICD-10), as well as guidelines and statements from sleep expert societies.
Based on the identified approaches, target systems are designed, based on previous work using the research imaging management system XNAT [9] and new developments in both biomedical data sharing and clinical decision support.
Results
Depending on the different targets, we defined data-driven solution approaches, including the primary data types that need to be made FAIR and available for the implementation of the solution. They are summarized in Table 1 [Tab. 1].
Table 1: Proposed solutions within the Somnolink project for the addressed targets, relevant data types, and current status of the conceptualization and implementation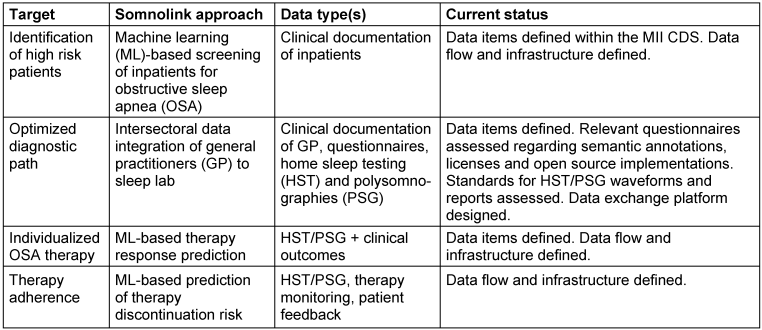
While target (b) – optimized diagnostic path – is addressed by improved data sharing between all healthcare actors that participate in the diagnostic processes, the other targets are addressed by developing ML-based prediction models for clinical decision support. In the following, we describe the required data types, existing standards, gaps and planned actions for the different targets.
Identification of high risk patients
To allow wide applicability, we have selected the data items from the base modules of the MII-CDS, (cf. Table 2 [Tab. 2]), except body weight and height that are only available in the extension module intensive care. The Body Mass Index, calculated from body height and body weight, is known as a relevant risk factor for OSA. As body weight and height are often assessed in the physical examination, we assume that these items are available although currently not defined in a base profile. The distinction between base and extension modules is relevant, because the Data Integration Centers of all University Medical Centers in Germany must provide the base modules, but can choose on a subset of extension modules.
Table 2: Data items identified as potential screening parameters.
To ensure wide availability, we tried to focus on base modules of the MI-CDS, except vital signs defined in the module Intensive Care. 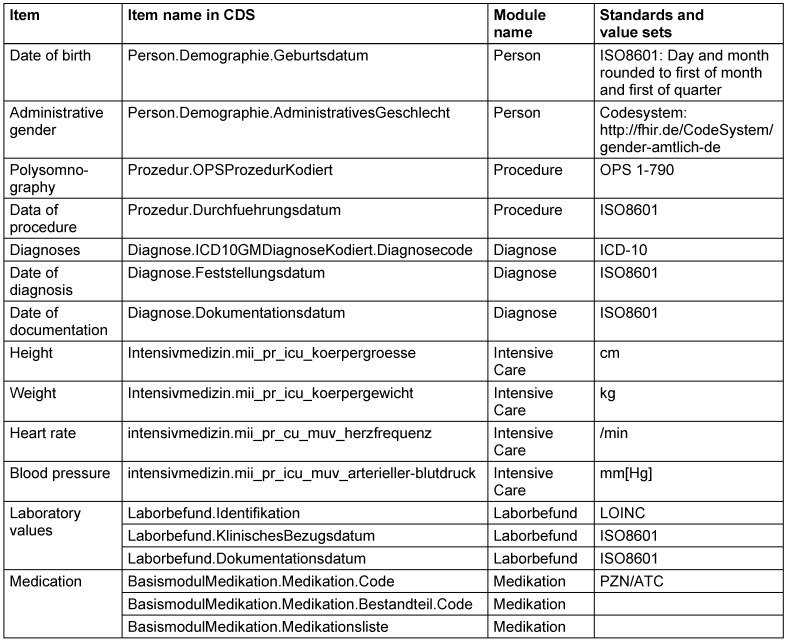
The item polysomnography is not required as a parameter for the model, but allows for validation, as OSA is reliably detected in a PSG.
A preliminary model is trained on a data set of one site, as this data was locally available through the state’s health data usage act. It includes also items that are currently not defined within the MII-CDS, i.e. the PSG and further sleep assessments for labeling the training data. The data flow for the development of the full ML-method based on retrospective big data analysis from all sites, is enabled from the sites to a central transfer office through the data sharing framework of the MII [10]. The data is stored centrally in production Extensible Neuroimaging Archive Toolkit (XNAT) instance hosted by the GWDG. The instance is connected to Jupyter notebooks for interactive preparatory data visualization and preparation, and to GWDG’s secure HPC workflow to reach a secure infrastructure capable for large scale ML training of sensitive data [11]. The outline of the data exchange and processing pipeline is given in Figure 2 [Fig. 2].
Figure 2: Data flow for data model training based on retrospective data received by the data integration centers (DIC). 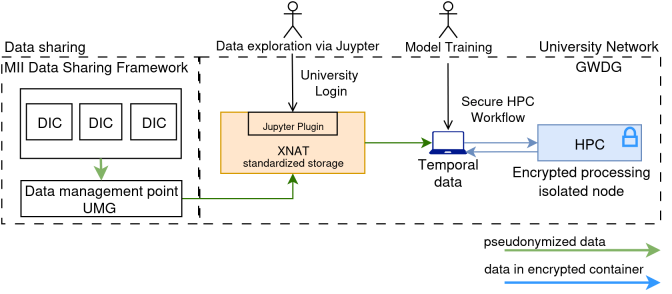
HPC: high performance computing, XNAT: Extensible Neuroimaging Archive Toolkit, MII: Medizininformatik-Initiative (German Medical Informatics Initiative), GWDG: Gesellschaft für wissenschaftliche Datenverarbeitung Göttingen (academic computer center Göttingen).
Optimized diagnostic path
As described above, the diagnostic path encompasses 4 steps, starting with a sleep assessment at the GP, diagnostic HST in the ambulatory setting and conditional diagnostic PSG and therapy initiation with PSG. The participating clinicians have prioritized the availability of the HTS waveforms, annotations and respective reports before therapy initiation.
The de-facto interoperability standard for biosignal waveforms is the European Data Format (EDF) [9]. It is compatible to the IEEE 11073 archive format, but has very limited metadata. EDF+ allows for the inclusion of annotations, but is rarely implemented by device vendors, that typically use proprietary xml-schema for annotations. We aim to transform the waveform data to DICOM as the final format. This allows for seamless integration in existing DICOM-based Picture Archiving and Communication Systems. However, each individual waveform is stored as a separate study; and all annotations that are not automatically generated during the recording is to be stored in a structured report according to the respective Supplement 239 of the DICOM specification [12]. Therefore data transfer will always encompass several individual DICOM objects, which is more complex to handle than an EDF-file.
Semantic codes in DICOM are typically inherited by the IEEE 11073 standard, with a license to use them in the context of DICOM. The defined annotations encompass sleep stages wake and Rapid Eye Movement (REM), as well as typical patterns in sleep recordings and apneas. Interestingly, it does not encompass the Non-REM sleep stages N1–N3 that are therefore defined in DICOM.
The content of a sleep report is defined by the American Association of Sleep Medicine (AASM), but there is no healthcare data format defined. Semantic codes for most items of a respiratory sleep report are defined as LOINC codes. For some reason, both the Apnea Index and the Hypopnea Index are defined in LOINC, while the typically reported Apnea-Hypopnea Index is only defined in SNOMED CT. The absence of an established standard for sleep assessments has led to the initiation of a sleep module as part of the MII-CDS.
Besides the biosignal recordings, questionnaires play a major role in the assessment of sleep quality and sleep disorders. The scores of the mainly used questionnaires, the Epworth Sleepiness Scale, the Pittsburgh Sleep Quality Index or the Insomnia Severity Index are coded in SNOMED CT. However, the individual questions and answers do not have semantic codes, except the Berlin Questionnaire, that is semantically annotated in detail with LOINC (cf. https://forms.loinc.org/62636-6). This is in line with an initiative to code questionnaires with LOINC [13]. Table 3 [Tab. 3] shows the most important data types in sleep diagnosis and therapy.
Table 3: Medical information assessed during OSA diagnosis and respective semantic standards.
While sensor-related information is related to DICOM and the IEEE 10073 standard, sleep parameters are available in LOINC. Diagnostic scores are typically found in SNOMED CT, while questionnaires could be coded in LOINC, but sleep-related questionnaires are rarely found. 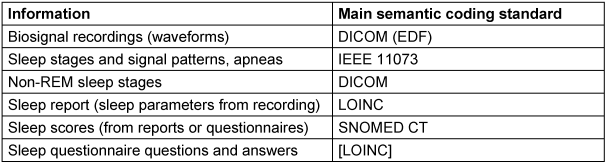
The data flow for the intersectoral data sharing is more complex, as identifying data is required for the diagnostic process. Based on the solutions for intersectoral health data sharing in the care context developed within the digital progress hubs of the MII, a central hub is currently implemented at the computer center to host questionnaires, clinical data from the electronic health record and HST/PSG biosignals, annotations and reports using the above mentioned healthcare standards. The main system for the exchange of waveforms is again XNAT (cf. Figure 2 [Fig. 2]), but connected to an additional FHIR server. Currently our practice partners test export from their local HTS-systems, upload to the Somnolink hub, webbased interactive visualization but also import in other vendor software. As the native XNAT user interface is quite We aim to make all data sharing compatible with the interoperability standards of the German healthcare system, for example employing the Matrix protocol for end-to-end encryption, to allow for smooth technical transfer into practice [14].
Individualized OSA therapy and therapy adherence
Both targets are closely related and are here presented together. They may influence each other. Optimized choice of therapy may improve therapy adherence, on the other hand may low adherence lead to a reexamination of the therapy choice. Both targets are addressed by clinical decision support systems. A comprehensive harmonized data item list has been consented, encompassing the dataset shown in Table 2 [Tab. 2], the sleep data included into the MII-CDS modules as well as further questionnaires, HST/PSG, therapy monitoring, sleep assessments and sociodemographic information. The models are pretrained with retrospective data available from the participating sleep labs and therapy monitoring systems. We are aware that the data will be incomplete due to the different patient collectives and primary specialty of the clinics where the sleep lab is located. Therefore a prospective study is currently planned including data collection during diagnosis. As the clinical decision support systems fall under the Medical Device Regulation, the study is designed as “other clinical investigation” according to article 82. The required documentation is currently compiled. The data flow for the model training on retrospective data is shown in Figure 2 [Fig. 2]. The data collection for the prospective study is realized through the Somnolink hub, with waveform data stored on XNAT and structured health data including questionnaires stored in the FHIR server.
Discussion and conclusion
Sleep medicine is a multidisciplinary specialty, and healthcare standards typically address only parts of the full data spectrum, including technical standards for biosignal recordings. However, most of the relevant data items and measures are covered by one of the semantic standards, allowing for semantic interoperability. Recent achievements in the German digital health data infrastructures for research and healthcare are promising to accelerate the project progress. However, regulatory requirements set by the European Medical Device Regulation, the AI Act and the Cyberresiliance Act are challenging not only for transfer into practice, but already in conducting research within the healthcare setting. Nevertheless, we are optimistic that the current engagement towards the European Health Data space will help to simplify these challenges.
Notes
Authors’ ORCIDs
- Dagmar Krefting: 0000-0002-7238-5339
- Michael Arzt: 0000-0002-4408-722X
- Moritz Brandt: 0000-0001-7686-7485
- Miriam Goldammer: 0000-0003-2126-290X
- Franz Ehrlich: 0000-0001-5682-9991
- Joachim T. Maurer: 0000-0003-2308-7041
- Thomas Penzel: 0000-0002-4304-0112
- Tony Sehr: 0000-0002-3156-0778
- Philip Zaschke: 0009-0001-5819-4309
- Christoph Schöbel: 0000-0002-0443-2973
Competing interests
The authors declare that they have no competing interests.
References
[1] Lim DC, Najafi A, Afifi L, Bassetti C, Buysse DJ, Han F, Högl B, Melaku YA, Morin CM, Pack AI, Poyares D, Somers VK, Eastwood PR, Zee PC, Jackson CL; World Sleep Society Global Sleep Health Taskforce. The need to promote sleep health in public health agendas across the globe. Lancet Public Health. 2023 Oct;8(10):e820-e826. DOI: 10.1016/S2468-2667(23)00182-2[2] Benjafield AV, Ayas NT, Eastwood PR, Heinzer R, Ip MSM, Morrell MJ, Nunez CM, Patel SR, Penzel T, Pépin JL, Peppard PE, Sinha S, Tufik S, Valentine K, Malhotra A. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med. 2019 Aug;7(8):687-98. DOI: 10.1016/S2213-2600(19)30198-5
[3] Chang JL, Goldberg AN, Alt JA, Mohammed A, Ashbrook L, Auckley D, et al. International Consensus Statement on Obstructive Sleep Apnea. Int Forum Allergy Rhinol. 2023 Jul;13(7):1061-482. DOI: 10.1002/alr.23079
[4] Askland K, Wright L, Wozniak DR, Emmanuel T, Caston J, Smith I. Educational, supportive and behavioural interventions to improve usage of continuous positive airway pressure machines in adults with obstructive sleep apnoea. Cochrane Database Syst Rev. 2020 Apr;4(4):CD007736. DOI: 10.1002/14651858.CD007736.pub3
[5] Semler SC, Boeker M, Eils R, Krefting D, Loeffler M, Bussmann J, Wissing F, Prokosch HU. Die Medizininformatik-Initiative im Überblick – Aufbau einer Gesundheitsforschungsdateninfrastruktur in Deutschland [The Medical Informatics Initiative at a glance-establishing a health research data infrastructure in Germany]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2024 Jun;67(6):616-28. DOI: 10.1007/s00103-024-03887-5
[6] Gesundheitsdatennutzungsgesetz (GDNG). BGBl Teil I. 2024 Mar 25;2024(102).
[7] Gesetz zur Beschleunigung der Digitalisierung des Gesundheitswesens (Digital-Gesetz – DigiG). BGBl Teil I. 2024 Mar 25;2024(101).
[8] Krefting D, Arzt M, Maurer JT, Penzel T, Prasser F, Sedlmayr M, Schöbel C. Sleep apnea healthcare management in dynamically changing times. Unlocking the potential of digitalization for better care of obstructive sleep apnea – in Germany and beyond. Somnologie. 2023 Oct 25;27:248-54. DOI: 10.1007/s11818-023-00428-1
[9] Beier M, Jansen C, Mayer G, Penzel T, Rodenbeck A, Siewert R, Witt M, Wu J, Krefting D. Multicenter data sharing for collaboration in sleep medicine. Future Generation Computer Systems. 2017 Feb;67:466-80. DOI: 10.1016/j.future.2016.03.025
[10] Hund H, Wettstein R, Kurscheidt M, Schweizer ST, Zilske C, Fegeler C. Interoperability Is a Process – The Data Sharing Framework. In: Bichel-Findlay J, Otero P, Scott P, Huesing E, editors. MEDINFO 2023 – The Future Is Accessible. Proceedings of the 19th World Congress on Medical and Health Informatics. IOS Press; 2024. p. 28-32. (Studies in Health Technology and Informatics; 310). DOI: 10.3233/SHTI230921
[11] Nolte H, Spicher N, Russel A, Ehlers T, Krey S, Krefting D, Kunkel J. Secure HPC: A workflow providing a secure partition on an HPC system. Future Generation Computer Systems. 2023 Apr 1;141:677-91. DOI: 10.1016/j.future.2022.12.019
[12] DICOM Standards Committee, Working Group 6. Digital Imaging and Communications in Medicine (DICOM). Supplement 239: Waveform Annotation SR. 2024. Available from: https://www.dicomstandard.org/News-dir/ftsup/docs/sups/sup239.pdf
[13] Vreeman DJ, McDonald CJ, Huff SM. Representing Patient Assessments in LOINC®. AMIA Annu Symp Proc. 2010 Nov 13;2010:832-6.
[14] The Matrix.org Foundation. Matrix. GitHub; [cited 2025 Jan 7]. Available from: https://github.com/matrix-org




