FAIVOR – a push-button system for AI validation within the hospital
Daniël Slob 1Ekaterina Akhmad 2
Jesus Garcia González 2
Saba Amiri 2
Vedran Kasalica 2
Sonja Georgievska 2
Ananya Choudhury 1
Aiara Lobo Gomes 1
Andre Dekker 1
Johan van Soest 1,3
1 Department of Radiation Oncology (Maastro), GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Centre+, Maastricht, The Netherlands
2 Netherlands eScience Center, Amsterdam, The Netherlands
3 Brightlands Institute for Smart Society (BISS), Faculty of Science and Engineering, Maastricht University, Heerlen, The Netherlands
Abstract
One notable challenge for pretrained medical AI models is their validation on new, unseen data. Developers train a model on a limited patient population and users must ensure that they deploy it on a clinical population without any statistical differences. The FAIVOR-tool aims to address these matters by proposing a robust system that enables the evaluation and adaptation of pretrained medical AI systems to new datasets. The tool includes the approach for containerization of AI models, an AI model repository and a tool to fetch and evaluate AI models on the local datasets. These objectives ensure responsible, transparent, and adaptable use of pretrained models in diverse clinical settings.
Keywords
artificial intelligence, medicine, evaluation, adaptation, deployment
1 Introduction
As artificial intelligence (AI) systems continue to gain importance in healthcare, ensuring responsible deployment into clinical practice is crucial. One important aspect of deployment is the assessment of the system’s capability to achieve set objectives on new, unseen data (hereafter referred to as “runtime data”). This process of local validation, however, is a privacy-sensitive matter that requires measurements to ensure data privacy is preserved on heterogeneous model architectures. Our FAIVOR tool is designed to tackle these challenges by facilitating local validation with the following considerations in mind: (1) strict data privacy compliance, (2) model-agnostic adaptability to various platforms and (3) robust evaluation beyond standard performance metrics.
To meet these challenges, the FAIVOR tool is developed with the following objectives: (1) containerization of AI models which can be downloaded inside the healthcare organisation, (2) a repository to register, search and find AI models, with references to the containerized AI models, and (3) a tool which fetches the AI models from the registry and evaluates the model on a local dataset. These objectives ensure responsible, transparent, and adaptable use of pretrained models in diverse clinical settings.
2 Methods
2.1 Requirements for local validation of AI models
Descriptive statistics of all input features and a patient cohort should be reported to assess cohort similarity. Statistical performance of the AI model should be evaluated based on the needs of the respective clinical context [1], [2], [3] and translated to essential metrics [4], [5], specified in model metadata [4], [6]. The documentation should report all requirements for input data, applicability criteria, essential metrics, and results of previous evaluations that could be considered to assess the local validation.
For reproducibility of testing, the local validation should be complemented with adequate data registration. Therefore, the complete description of AI models and validation results in different clinical settings should be stored in a general repository of AI models while providing the model in executable format.
2.2 Preparation of AI models
A commonly used tool for model containerization is Docker. A Docker image can be built including specified dependencies, versions and functions to calculate the actual prediction (e.g. weight coefficients in case of linear regression models). To interact with the model, a standardised REST API will be embedded in the Docker container.
The model validation requires access to the model description. Halilaj et al. presented an open-source repository for clinical prediction models [7]. However, model reports are limited to only storing model coefficients and no validation possibilities were provided. The repository for our project should provide a comprehensive description using FAIR principles, applied to AI models, to enable interoperability of the AI model.
2.3 Tool to fetch the AI models from the registry
A GUI-based application will retrieve models from the FAIR registry, run local validations, generate statistics, and publish results. It simplifies model evaluation and helps identify performance trends across hospitals and over time.
3 Results
The FAIVOR tool is an on-premises system for evaluating and adapting pretrained medical machine learning models to new datasets. The tool encompasses the following architecture (Figure 1 [Fig. 1]) (https://github.com/MaastrichtU-BISS/FAIRmodels-validator).
Figure 1: Architecture of the FAIVOR tool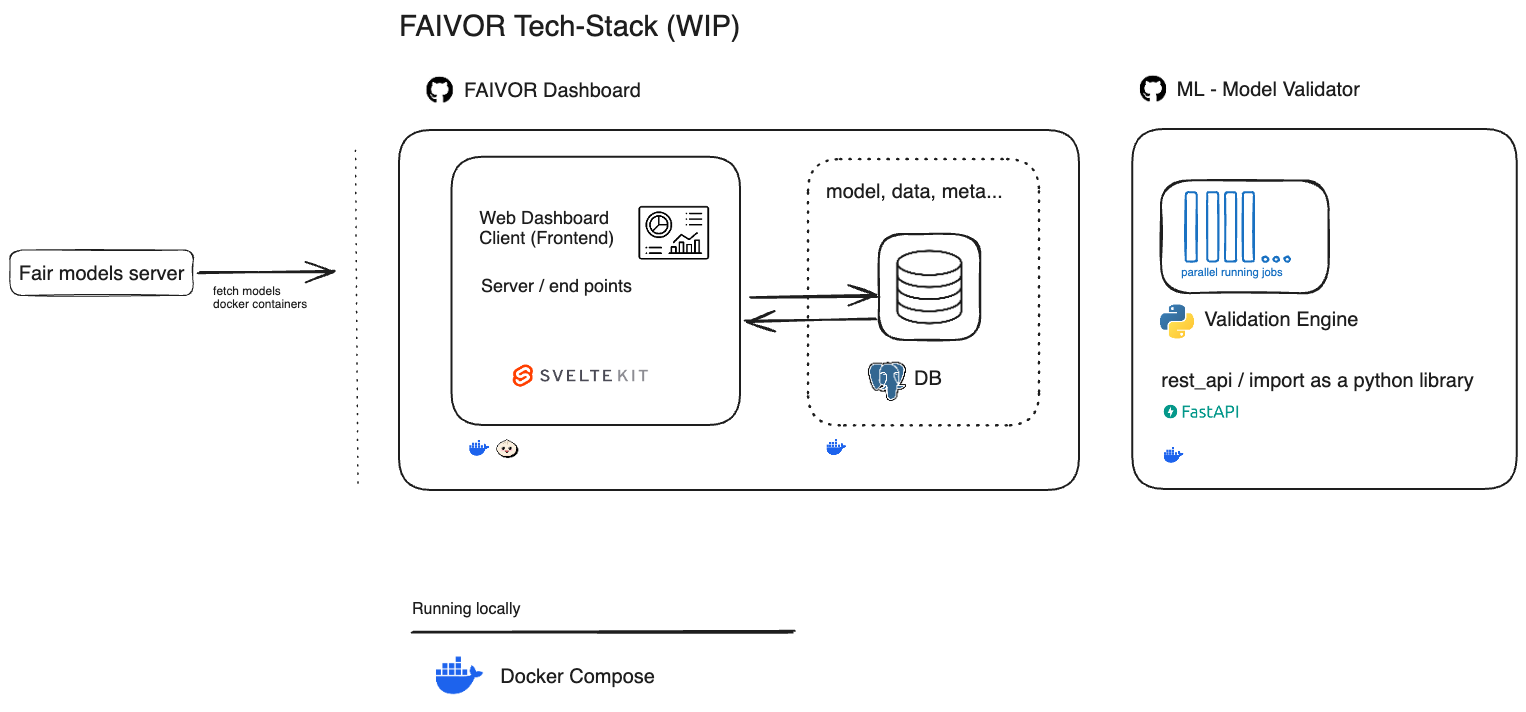
The AI model described in Stiphout et al. [8] serves as a use case to illustrate the FAIVOR tool architecture. Before starting their validation job, the developers of [8] had uploaded their AI model – with corresponding metadata – to the model repository (https://v2.fairmodels.org/instance/3f400afb-df5e-4798-ad50-0687dd439d9b). The repository guided the developers to generate FAIR metadata for their model. The model was then packaged in Docker images, following instructions on https://github.com/MaastrichtU-BISS/FAIRmodels-model-package/. After requirements were met, the URI (URL) of the model metadata [8] was ready to be fetched from the repository. The newly created Docker image containing the AI system was then pulled from the specified location on the model metadata and used for validation.
An intuitive GUI guided the developers in setting the preliminary grounds for their validation job. The GUI included features that allowed the developers to keep track of the validation job(s) status(es), document the validation job using a standardized format, select from a range of predetermined metrics based on [4], [5] (e.g. PPV, NPV), and calculate (I) summary statistics for continuous and categorical features and (II) performance evaluation metrics.
4 Discussion and conclusions
A robust, privacy-preserving tool is developed to evaluate and adapt pretrained medical machine learning models to new datasets. The FAIVOR tool shows similarities to existing tools such as MONAI [9], MLflow and EvalAI [10]. It differentiates by supporting FAIR metadata generation, focusing on the clinical context specifically and facilitating local validation for models that use tabular data as input. Limitations, however, should be acknowledged as the tool is still under development. At this stage, the tool only supports AI systems trained on tabular data; medical images are not available to be used. Future work includes developing the ability to upload evaluation results into the registry and options to compare validation jobs to facilitate on-premises continuous monitoring.
Notes
Author’s ORCID
Daniël Slob: 0009-0002-2084-5660
Competing interests
The authors declare that they have no competing interests.
References
[1] Bibb A, Dreyer K, Stibolt R, Agarwal S, Coombs L, Treml L, Elkholy M, Brink L, Wald C. Evaluation and Real-World Performance Monitoring of Artificial Intelligence Models in Clinical Practice: Try It, Buy It, Check It. JACR. 2022 Nov;18(11):1489-96. DOI: 10.1016/j.jacr.2021.08.022[2] Altman DG, Vergouwe Y, Royston P, Moons KG. Prognosis and prognostic research: validating a prognostic model. BMJ. 2009 May 28;338:b605. DOI: 10.1136/bmj.b605
[3] Scott I, Carter S, Coiera E. Clinician checklist for assessing suitability of machine learning applications in healthcare. BMJ Health Care Inform. 2021 Feb;28(1):e100251. DOI: 10.1136/bmjhci-2020-100251
[4] Binuya MAE, Engelhardt EG, Schats W, Schmidt MK, Steyerberg EW. Methodological guidance for the evaluation and updating of clinical prediction models: a systematic review. BMC Med Res Methodol. 2022 Dec 12;22(1):316. DOI: 10.1186/s12874-022-01801-8
[5] Cabitza F, Campagner A. The need to separate the wheat from the chaff in medical informatics: Introducing a comprehensive checklist for the (self)-assessment of medical AI studies. Int J Med Inform. 2021 Sep;153:104510. DOI: 10.1016/j.ijmedinf.2021.104510
[6] Tanguay W, Acar P, Fine B, Abdolell M, Gong B, Cadrin-Chênevert A, Chartrand-Lefebvre C, Chalaoui J, Gorgos A, Chin AS, Prénovault J, Guilbert F, Létourneau-Guillon L, Chong J, Tang A. Assessment of Radiology Artificial Intelligence Software: A Validation and Evaluation Framework. Can Assoc Radiol J. 2023 May;74(2):326-33. DOI: 10.1177/08465371221135760
[7] Halilaj I, Oberije C, Chatterjee A, van Wijk Y, Rad NM, Galganebanduge P, Lavrova E, Primakov S, Widaatalla Y, Wind A, Lambin P. Open Source Repository and Online Calculator of Prediction Models for Diagnosis and Prognosis in Oncology. Biomedicines. 2022 Oct 23;10(11):2679. DOI: 10.3390/biomedicines10112679
[8] van Stiphout RG, Lammering G, Buijsen J, Janssen MH, Gambacorta MA, Slagmolen P, Lambrecht M, Rubello D, Gava M, Giordano A, Postma EO, Haustermans K, Capirci C, Valentini V, Lambin P. Development and external validation of a predictive model for pathological complete response of rectal cancer patients including sequential PET-CT imaging. Radiother Oncol. 2011 Jan;98(1):126-33. DOI: 10.1016/j.radonc.2010.12.002
[9] Cardoso MJ, Li W, Brown R, Ma N, Kerfoot E, Wang Y, Murrey B, et al. MONAI: An open-source framework for deep learning in healthcare [Preprint]. arXiv. 2022 Nov 4. DOI: 10.48550/arXiv.2211.02701
[10] Yadav D, Jain R, Agrawal H, Chattopadhyay P, Singh T, Jain A, Singh SB, Lee S, Batra D. EvalAI: Towards Better Evaluation Systems for AI Agents [Preprint]. arXiv. 2019 Feb 10. DOI: 10.48550/arXiv.1902.03570




