Molecular detection of phenol-soluble modulin-mec (PSM-mec) in Staphylococcus aureus clinical isolates from Federal Medical Center Birnin Kebbi, North-West, Nigeria
Isah Musa Maishanu 1,2Adeshina O. Gbonjubola 2
Hussaini Mujahid 2,3
Busayo O. Olayinka 2
1 Kebbi State University of Science and Technology Aliero, Nigeria
2 Ahmadu Bello University Zaria, Kaduna, Nigeria
3 Ummaru Musa Yaradua University Katsina, Nigeria
Abstract
Aim: This study was carried out to isolate and detect virulence genes associated with Staphylococcus (S.) aureus clinical isolates from the Federal Medical Center Birnin Kebbi, Nigeria.
Methods: To obtain S. aureus isolates, samples were taken from urine, sputum, blood and wound sources. S. aureus was phenotypically identified using Microgen staph ID system and PSM-mec and PVL genes were detected using polymerase chain reaction (PCR).
Results: A total of 48 non-duplicate S. aureus isolates were obtained (21 from wound swabs, 7 from blood, 15 from urine, and 5 from sputum). From the 14 S. aureus isolates examined by PCR, the most abundant gene was PSM-mec (42.8%), while the PVL was the least abundant with 21.4%.
Conclusion: Because it gives highly specific and accurate results, it is essential to use the PCR technique to detect S. aureus virulence determinants as well as PSM-mec and PVL as targets for antimicrobial agents.
Keywords
Staphylococcus aureus, phenol soluble modulin-mec, panton valentine leucocidin, virulence genes
Introduction
Being one of the “ESKAPE” organisms, Staphylococcus (S.) aureus poses an increasing hazard to human health due to its ability to produce a range of serious nosocomial infections [1]. The relationship among bacterial evolution, host factors, and virulence determinants has been the focus of recent clinical research [2], [3]. The virulence determinants found in Staphylococcus aureus comprise staphylokinase, hyaluronidase, lipase, nuclease, hemolysin, leukocidin, and invasive proteases. Leukocidin induces inflammatory reactions by disrupting skin, mucosal cells, and, among the host blood cells, leukocytes [4]. The genes that codes for leukocidin are PSM-mec, luk-F/-S-PV, lukE, lukM, and PSM-α. Due to the presence or lack of mobile genetic elements (MGEs), which are made up of genes encoding for toxins and other virulence factors, the organism’s potential for virulence varies greatly amongst isolates. Antibiotic resistance determinants and virulence factors proliferate as a result of the rich diversification of the naturally adapted mobile genetic element Staphylococcus cassette chromosome mec (SCCmec), which is responsible for the stable maintenance of the core genome environment. Unlike several other bacterial pathogens, which frequently depend on just one or a few toxins to cause illness, S. aureus generates an incredible variety of virulence factors. These comprise a wide range of protein and non-protein components that facilitate host colonization during infection, as well as an abundance of toxins and immune evasion mechanisms. Staphylococcal cassette chromosome mec (SCCmec) elements contain PSM-mec, as well as regulatory factors, recombinase genes, mecA, and other resistance genes [5], [6]. The sole known virulence determinant associated with these determinants is the PSM-mec. It has been discovered in S. aureus SCCmec types II, III, and VIIIK [7]. Multiple cell surface and secreted virulence factors mediate the pathogenesis of S. aureus. One such virulence factor is called Panton-Valentine leukocidin (PVL), an extracellular protein with dermonecrotic and leucocidal properties, which is expressed by the genes luk-S-PV and luk-F-PV. It is cytotoxic to macrophages, monocytes, and neutrophils in mammals. Community-acquired methicillin-susceptible S. aureus (CA-MSSA) and community-acquired MRSA (CA-MRSA) strains can both produce the toxin. In addition to causing SSTIs, PVL-positive strains have been connected to purpura fulminans, necrotizing pneumonia, bacteremia, and septic arthritis, among other serious diseases [8]. Detecting PSM-mec and PVL genes in S. aureus clinical isolates from the Federal Medical Centre Birnin Kebbi was the aim of the current investigation.
Materials and methods
Collection and authentication of bacterial isolates
A total of 120 presumptive staphylococcal isolates were collected from wound swabs, urine, blood and sputum. Following conventional microbiological protocols, the isolates were cultivated and identified using the Microgen Staph ID system (.Microgen bioproducts Ltd, UK).
DNA extraction
Each sample was grown for an entire night at 33°C for 24 hrs on Mueller-Hinton agar plates. After that, 3 ml of sterile lysogeny broth medium were used to placed each single bacterial colony (Oxoid, Hampshire, UK), which was then cultured for eight hours at 33°C with vigorous shaking. Next, DNA was extracted using the Hipure Bacterial DNA Kit (Magen, Guangzhou, China) in compliance with the manufacturer’s instructions.
Amplification of virulence genes using PCR
PCR was used to amplify PSM-mec and PVL, as previously reported by Jiang [9] in 14 isolates of S. aureus. Table 1 [Tab. 1] lists the target genes, primer sequences, and target segment of the PCR products. The PCR techniques were performed in a final volume of 25 µL of reaction mixture that contained 50 ng of genomic DNA, 20 pmol of each primer, and 12.5 µL of 2×Taq PCR Master Mix (Tiangen Biotech, China: 0.1 U of Taq polymerase/µL, 0.5 mM dNTP each, 20 mM Tris-HCl/pH 8.3, 100 mM KCl, 3 mM MgCl2). The denaturation process was completed in 3 minutes at 94°C, followed by 30 cycles of 30 seconds each at 94°C for denaturation, 30 seconds of annealing at 55°C 1 minute of primer extension at 72°C, and 5 minutes of final extension at 72°C.
Table 1: The primers used and their nucleotide sequences
Statistical analysis
The statistical software for social sciences (SPSS) version 21 was used to analyze the data. The analysis employed descriptive statistics such as percentages.
Results
Forty-eight (48) S. aureus isolates were obtained during the course of a 6-month study period. The majority of S. aureus isolates were from wound (43.7%), urine (31.25%) and blood (12.5%) while sputum was (10.4%). A breakdown of the prevalence of (17.5%) in wound samples was recorded as shown in (Table 2 [Tab. 2]).
Table 2: Prevalence of Staphylococcus aureus isolates by specimen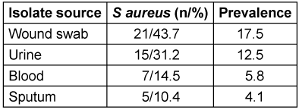
The prevalence of PVL and PSM-mec genes in 14 of the S. aureus isolates was 21.4% (3/14) and 42.8% (6/14), respectively. The highest proportion of PSM-mec genes was detected in the S. aureus isolates from wound swabs, followed by urine, while PVL genes were also detected in mostly in wound swabs, followed by blood (Figure 1 [Fig. 1]).
Figure 1: Percentage of virulence genes in S aureus from various clinical specimens. (PVL=panton valentine leucocidin, PSM-mec=phenol soluble modulin-mec)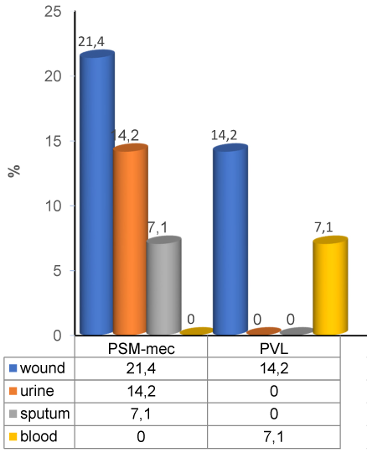
Discussion
In the community, hospital, and environmental settings, virulence factors are an essential component of pathogenic invasion that results in staphylococcal infection. Global reports indicate that S. aureus possesses a rich diversity of virulence-associated genes [10], [11]. The virulence analysis was focused on detecting PVL and PSM-mec genes. Research has shown that the prevalence of PVL in S. aureus isolates derived from clinical specimens varies greatly between nations, with prevalences as high as 57% observed in isolates from west African countries and 9.7% in England [12], [13]. We investigated the prevalence of PVL genes in 14 clinical samples that tested positive for S. aureus. A prevalence of 33.0% was detected out of the 14 S. aureus isolates that underwent investigation for the PVL gene. This is substantially greater than what was found in some studies conducted in Nigeria, with 10.7% [14], 11.2% [15], [16] and 13% [17], but was similar to others, e. g, in Jos Nigeria with 31.3% [18], 34% in Southwest Nigeria [19] and 39.35% at Obafemi Awolowo University [20]. In contrast, our results were lower than in Maiduguri, Nigeria, which had 52.1% [21], Gambiya 61% [22], Sudan 58% [23], Iran 56% [24] and India 61% [25].
The virulence factor PSM-mec belongs to the class of amphipathic, alpha-helical peptide poisons known as phenol-soluble modulins (PSM). All known PSMs are core-genome encoded, with the exception of the PSM-mec, which is present in specific subtypes of SCCmec methicillin-resistant mobile genetic elements, discovered in methicillin-resistant S. aureus. According to Wang et al. [25], the alpha-type phenol-soluble modulins (PSMs), which are novel cytolytic peptides, are encoded in an operon present in every strain of S. aureus that has been sequenced. Recent studies suggest that PSM synthesis is inhibited by the transcription and translation products of PSM-mec, which are present in the HA-MRSA mobile genetic elements SCCmec-II and -III [26]. Queck et al. [27] discovered that PSM-mec had a positive impact on the pathogenicity of the HA-MRSA strain MSA890. Also, the translational product of PSM-mec was found to be more prevalent than that of other PSMs. In the present study, only 42.8% of the 14 MRSA isolates that underwent PCR analysis carried the PSM-mec gene. The majority of HA-MRSA carries the PSM-mec gene, which is present in type-II and type-III SCCmec and regulates S. aureus pathogenicity [28]. The S. aureus PSM-mec gene, which is necessary for MRSA colonization and pathogenesis, is still not well understood. Given the essential roles of the PVL and PSM-mec genes, these virulence factors may be potential candidates for consideration in vaccines that combat MRSA strains.
Conclusions
S. aureus isolated from clinical samples in Nigeria possessed the PSM-mec and PVL genes. An understanding of the links between virulence and resistance would help to lessen the impact of S. aureus infections, given the high prevalence of infections produced by this pathogen and its significance in human medicine. To obtain deeper insight into the various virulence and resistance mechanisms employed by this pathogen and their interactions, additional research is required.
Notes
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Ethical clearance was obtained from the ethical committee of Federal Medical Center Birnin Kebbi, Nigeria to enable collection of Staphylococcal clinical isolates from medical microbiology unit of the hospital.
Funding
None.
Acknowledgments
We are very grateful to members of staff of Microbiology unit, Federal Medical Center Birnin Kebbi and that of Central Research Laboratory, Department of Veterinary Microbiology Usman Dan Fodio University Sokoto for their cooperation and assistance during the study.
References
[1] Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015 Aug;14(8):529-42. DOI: 10.1038/nrd4572[2] Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, Khoon LY, Aziz MN, Hamat RA, Othman N, Chong PP, van Belkum A, Ghasemzadeh-Moghaddam H, Neela V. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol. 2010 Mar;48(3):867-72. DOI: 10.1128/JCM.01112-09
[3] Xu S, Fu Z, Zhou Y, Liu Y, Xu X, Wang M. Mutations of the Transporter Proteins GlpT and UhpT Confer Fosfomycin Resistance in. Front Microbiol. 2017;8:914. DOI: 10.3389/fmicb.2017.00914
[4] Boan P, Tan HL, Pearson J, Coombs G, Heath CH, Robinson JO. Epidemiological, clinical, outcome and antibiotic susceptibility differences between PVL positive and PVL negative Staphylococcus aureus infections in Western Australia: a case control study. BMC Infect Dis. 2015 Jan;15:10. DOI: 10.1186/s12879-014-0742-6
[5] Chatterjee SS, Chen L, Joo HS, Cheung GY, Kreiswirth BN, Otto M. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(12):e28781. DOI: 10.1371/journal.pone.0028781
[6] Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol. 2013 Oct;11(10):667-73. DOI: 10.1038/nrmicro3110
[7] Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 2013;9(4):e1003269. DOI: 10.1371/journal.ppat.1003269
[8] He C, Xu S, Zhao H, Hu F, Xu X, Jin S, Yang H, Gong F, Liu Q. Leukotoxin and pyrogenic toxin Superantigen gene backgrounds in bloodstream and wound Staphylococcus aureus isolates from eastern region of China. BMC Infect Dis. 2018 Aug;18(1):395. DOI: 10.1186/s12879-018-3297-0
[9] Jiang B, Yin S, You B, Gong Y, Huang G, Yang Z, Zhang Y, Chen Y, Chen J, Yuan Z, Hu X, Peng Y. Antimicrobial resistance and virulence genes profiling of methicillin-resistant Staphylococcus aureus isolates in a burn center: A 5-year study. Microb Pathog. 2018 Jan;114:176-9. DOI: 10.1016/j.micpath.2017.11.020
[10] Sabouni F, Mahmoudi S, Bahador A, Pourakbari B, Sadeghi RH, Ashtiani MT, Nikmanesh B, Mamishi S. Virulence Factors of Staphylococcus aureus Isolates in an Iranian Referral Children's Hospital. Osong Public Health Res Perspect. 2014 Apr;5(2):96-100. DOI: 10.1016/j.phrp.2014.03.002
[11] 13 Chen X, Wu Z, Zhou Y, Zhu J, Li K, Shao H, et al. Molecular and virulence characteristics of methicillin-resistant Staphylococcus aureus in burn patients. Front Lab Med. 2017;1(1):43-7. DOI: 10.1016/j.flm.2017.02.010
[12] Breurec S, Fall C, Pouillot R, Boisier P, Brisse S, Diene-Sarr F, Djibo S, Etienne J, Fonkoua MC, Perrier-Gros-Claude JD, Ramarokoto CE, Randrianirina F, Thiberge JM, Zriouil SB; Working Group on Staphylococcus aureus InfectionsGarin B, Laurent F. Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin Microbiol Infect. 2011 Apr;17(4):633-9. DOI: 10.1111/j.1469-0691.2010.03320.x
[13] Shallcross LJ, Williams K, Hopkins S, Aldridge RW, Johnson AM, Hayward AC. Panton-Valentine leukocidin associated staphylococcal disease: a cross-sectional study at a London hospital, England. Clin Microbiol Infect. 2010 Nov;16(11):1644-8. DOI: 10.1111/j.1469-0691.2010.03153.x
[14] O’Malley SM, Emele FE, Nwaokorie FO, Idika N, Umeizudike AK, Emeka-Nwabunnia I, Hanson BM, Nair R, Wardyn SE, Smith TC. Molecular typing of antibiotic-resistant Staphylococcus aureus in Nigeria. J Infect Public Health. 2015;8(2):187-93. DOI: 10.1016/j.jiph.2014.08.001
[15] Babatunde OJ, Ogundare AO, Adebolu TT. Antibacterial activities of Polyalthia longifolia leaf extracts on multiple antibiotic-resistant bacteria isolated from hospital fomites in Akure, Nigeria. Nusantara Biosci. 2023;15(2). DOI: 10.13057/nusbiosci/n150203
[16] Abdullahi IN, Issaoui R, Usman Y. Prevalence and genetic lineages of nasal colonization and urinary tract infection among people living with HIV/AIDS in Nigeria: A systematic review. IJID Reg. 2022 Sep;4:17-24. DOI: 10.1016/j.ijregi.2022.05.009
[17] Orji OL, Olayinka BO, Afolabi B, Ejikeugwu Chika P, Nwakaeze EA. Molecular detection of panton-valentine leucocidin (PVL) toxins in clinical isolates of Staphylococcus aureus from Maitama district hospital. J Med Microb Diagn. 2016; 5:3. DOI: 10.4172/2161-0703.1000240
[18] Essien UC, Boswihi SS, Agbakoba NR, Udo EE. Description of Methicillin-Susceptible Staphylococcus aureus Clonal Complex 30 Related to the Pandemic Phage Type 80/81 Isolated from Patients in Three Tertiary Hospitals in Jos, North Central Nigeria. Med Princ Pract. 2022;31(3):269-75. DOI: 10.1159/000524755
[19] Alli OA, Ogbolu DO, Shittu AO, Okorie AN, Akinola JO, Daniel JB. Association of virulence genes with mecA gene in Staphylococcus aureus isolates from Tertiary Hospitals in Nigeria. Indian J Pathol Microbiol. 2015;58(4):464-71. DOI: 10.4103/0377-4929.168875
[20] Kolawole DO, Adeyanju A, Schaumburg F, Akinyoola AL, Lawal OO, Amusa YB, Köck R, Becker K. Characterization of colonizing Staphylococcus aureus isolated from surgical wards' patients in a Nigerian university hospital. PLoS One. 2013;8(7):e68721. DOI: 10.1371/journal.pone.0068721
[21] Okon KO, Uba A, Oyawoye OM, Yusuf IZ, Adesina OO. Prevalence and antibiotic susceptibility pattern of panton valentine leucocidin (PVL) positive Staphylococcus aureus strains from clinical specimens in Northeastern Nigeria. Sierra Leone J Biomed Res. 2012; 4(1):43-52.
[22] Darboe S, Dobreniecki S, Jarju S, Jallow M, Mohammed NI, Wathuo M, Ceesay B, Tweed S, Basu Roy R, Okomo U, Kwambana-Adams B, Antonio M, Bradbury RS, de Silva TI, Forrest K, Roca A, Lawal BJ, Nwakanma D, Secka O. Prevalence of Panton-Valentine Leukocidin (PVL) and Antimicrobial Resistance in Community-Acquired Clinical in an Urban Gambian Hospital: A 11-Year Period Retrospective Pilot Study. Front Cell Infect Microbiol. 2019;9:170. DOI: 10.3389/fcimb.2019.00170
[23] Osman NM, Alrayah IE, Mohamed YM, Erag AE, Eldirdery MM, Salih MA. Molecular study of panton-valentine leukocidin genes among Staphylococcus aureus clinical isolates in Khartoum State, Sudan. American J Microbiol Res. 2015; 3(3):107-11. DOI: 10.12691/ajmr-3-3-2
[24] Nourbakhsh V, Nourbakhsh F, Tajbakhsh E, Borooni S, Daneshmand D. Characterization of Staphylococcus aureus isolated from wound infectious in diabetes clinic of Hazrat Fatemeh Zahra (SA) hospital. Avicenna J Clin Microbiol Infect. 2018;5(3): 67-70. DOI: 10.34172/ajcmi.2018.13
[25] Patil NR, Ghorpade MV. Association of virulence factor (panton-valentine leukocidin) with meca gene in Staphylococcus aureus isolates in tertiary care hospital. Asian J Pharm Clin Res. 2018; 11(2): 113-6. DOI:10.22159/ajpcr.2018.v11i2.19080
[26] Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007 Dec;13(12):1510-4. DOI: 10.1038/nm1656
[27] Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, Deleo FR, Otto M. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009 Jul;5(7):e1000533. DOI: 10.1371/journal.ppat.1000533
[28] Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 2011 Feb;7(2):e1001267. DOI: 10.1371/journal.ppat.1001267
[29] Grace JU, Obaro S, Olayinka B, Onaolapo J, Hassan-Hanga F, Munir H et al. Staphylococcus epidermidis in Bloodstream Infection: The Clinical Implication in Paediatric Care. 2019. DOI: 10.13140/RG.2.2.26861.13289
[30] Le Thomas I, Mariani-Kurkdjian P, Collignon A, Gravet A, Clermont O, Brahimi N, Gaudelus J, Aujard Y, Navarro J, Beaufils F, Bingen E. Breast milk transmission of a Panton-Valentine leukocidin-producing Staphylococcus aureus strain causing infantile pneumonia. J Clin Microbiol. 2001 Feb;39(2):728-9. DOI: 10.1128/JCM.39.2.728-729.2001




